Complex models for the description of biological structures
- Introduction
- Biomolecular Structural Analysis
- Deformable Models for the Analysis of Cells and Subcellular Structures
- Analysis of Protoplasts: Description of Cell Anatomy and Similarity Measures for Complex Protein Patterns
- Automated measurement of hypocotyl growth rates of Arabidopsis seedlings
- Analysis of Organ Development On Sub-Cellular Resolution at the example of Arabidopsis Thaliana
- Analysis of Trichome Patterning and Leaf Anatomy of Arabidopsis Thaliana in 4D Confocal Datasets
Introduction
For various biological tasks it is important to have automated methods for the quantitative description of the phenotype of the specific organism in order to speed up or even allow experimental evaluations.
Thus an important focus lies on the development of models that are able to robustly describe the evolving biological anatomy ranging from the level of whole organisms to cell clusters, single cells and the subcellular level. We are primarily dealing with 3D(+time) but also 2D(+time) microscopic data.
Another important task in this context is the description of complex biological signals and patterns and their representation in meaningful anatomical coordinates obtained from the anatomical reference models.
Besides difficult properties of microscopic data imposed by imaging methods challenges are given by the spatio-temporal variations of biological anatomy at the different scales.
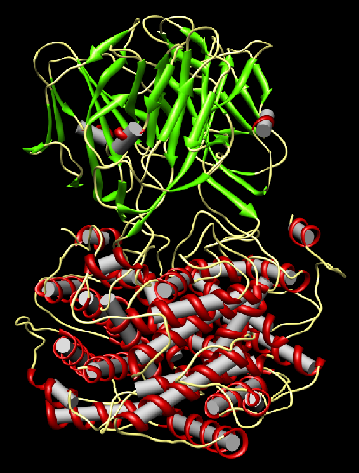
Biomolecular Structural Analysis
Members: Lingyu Ma, Prof. Dr.-Ing. Hans Burkhardt
Partners: Marco Reisert (Medical Physics, Department of Diagnostic Radiology, University Hospital Freiburg)
Introduction
Goals
Approach
|
|
Deformable Models for the Analysis of Cells and Subcellular Structures
Members: Margret Keuper, Junior-Prof. Dr. Olaf Ronneberger, Prof. Dr.-Ing. Hans Burkhardt
Partners: Jan Padeken,
Patrick Heun (Max-Planck Institute of Immunobiology)
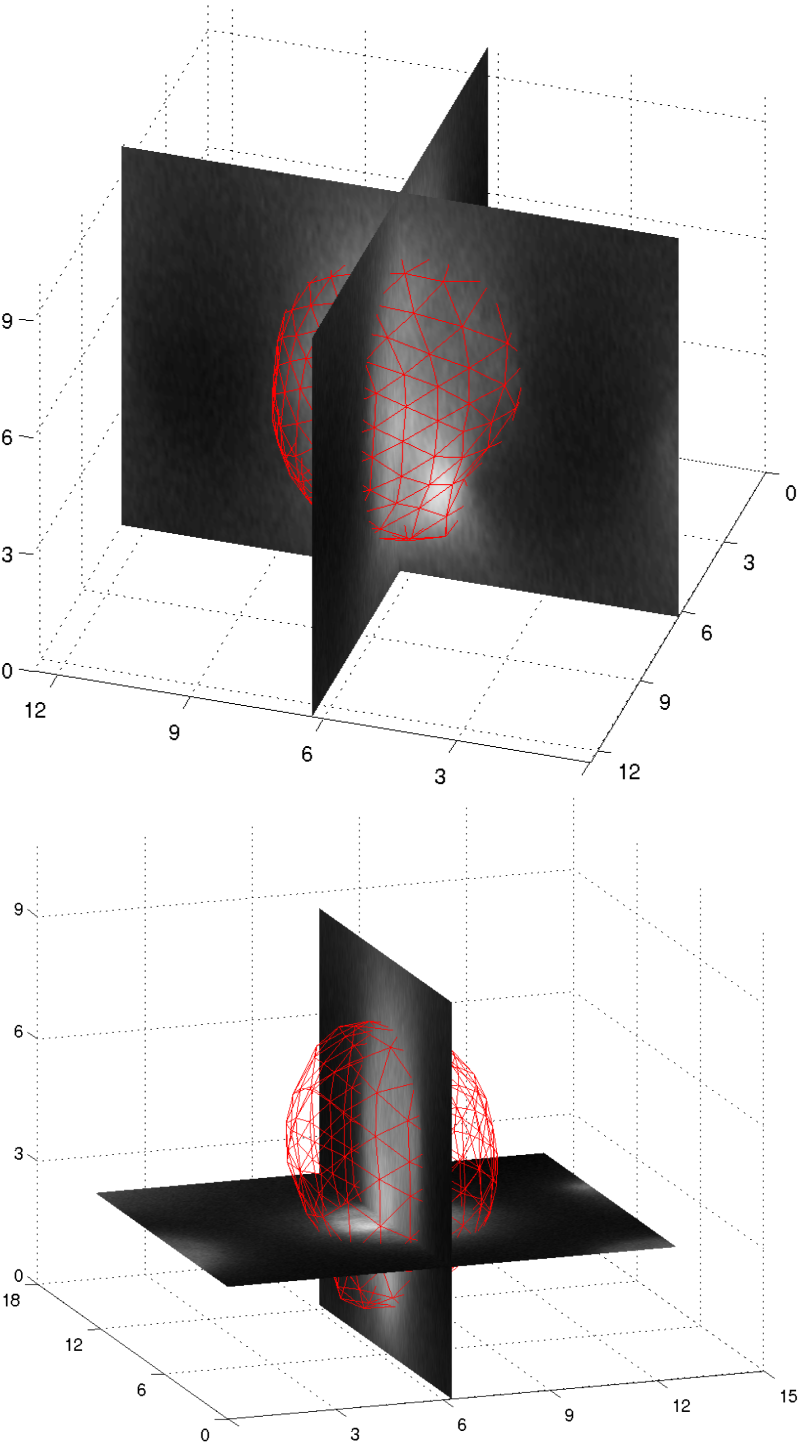
IntroductionFor the analysis of cellular and subcellular mechanisms, exact knowledge about the cell anatomy is needed. In this subproject, we are therefore developing methods to precisely describe the shapes and shape variations of 3D objects (cells, cell nuclei, nucleoli) in microscopic recordings. GoalsWe are aiming to develop a 3D deformable model framework that is able to decribe shapes and shape variations of different cell types from different organisms (drosophiola S2 cells, arabidopsis thaliana root cells, tobacco protoplasts). The models should be able to handle the noisy and blurred data from different microscopy techniches (from widefield fluorescence microscopy over spinning disk microscopy to confocal laser scanning microscopy) ApproachWe are combining methods to generate robust egde maps with 3D deformable surfaces. The segmentation consists of the following steps:
In step 1, the introduction of application specific user knowledge is necessary. In many cases, only the expert knows which structure he or she is looking for. This knowledge is learned from one or more training samples. In step 2, we are using parametric and non-parametric deformable surface models. Thus, we can introduce prior shape knowledge that facilitates the segmentation. The parametric segmentation at the same time yields a shape descriptor that can be used for the further description of the structure. AchievementsThe segmentation and modeling of cells and cell nuclei can be performed very robustly and despite the presence of noise and distortions, originating from the recording technique (CLSM and Widefield fluorescence microscopy) Publications
|
|
Analysis of Protoplasts: Description of Cell Anatomy and Similarity Measures for Complex Protein Patterns
Members: Robert Bensch, Margret Keuper, Henrik Skibbe, Junior-Prof. Dr. Olaf Ronneberger, Prof. Dr.-Ing. Hans Burkhardt
Partners: Karsten Voigt, Dr. Alexander Dovzhenko, Prof. Dr. Klaus Palme (Institute of Biology II, University of Freiburg)
IntroductionStudying protoplast cell development aims to increase the systems-understanding of molecular processes underlying the control of cell polarity, division, and the reprogramming [reference 1]. This will facilitate identification of key elements and gene networks responsible for totipotency in protoplasts. GoalsIn this context we pursue the goal to develop methods for a robust description of the temporally evolving 2D and 3D cell anatomy of growing and dividing protoplasts that build cell clusters. Moreover robust similarity measures for the description and comparison of complex 3D protein patterns that are invariant to imaging conditions and stainings are developed.
References
Publications
|
Automated measurement of hypocotyl growth rates of Arabidopsis seedlings
Members: Dr. Qing Wang, Junior-Prof. Dr. Olaf Ronneberger, Prof. Dr.-Ing. Hans Burkhardt
Partners: Cornelia Klose, Dr. Stefan Kircher, Prof. Dr. Eberhard Schäfer
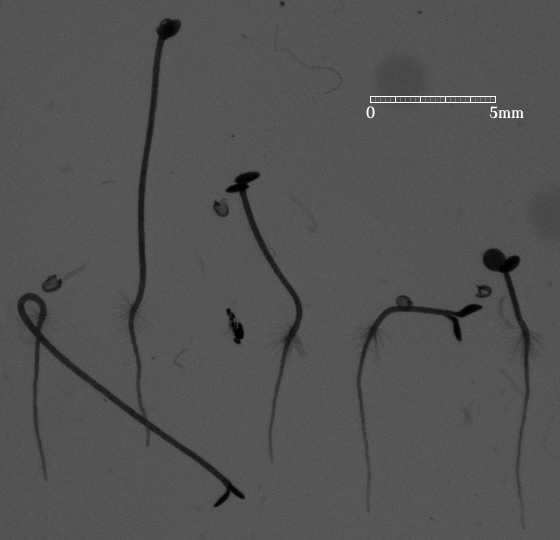
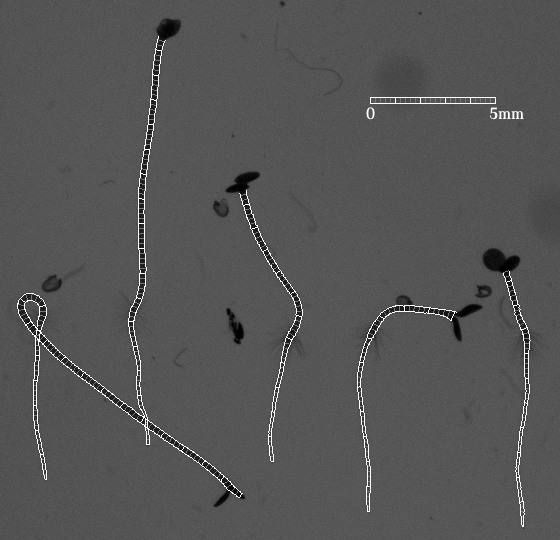
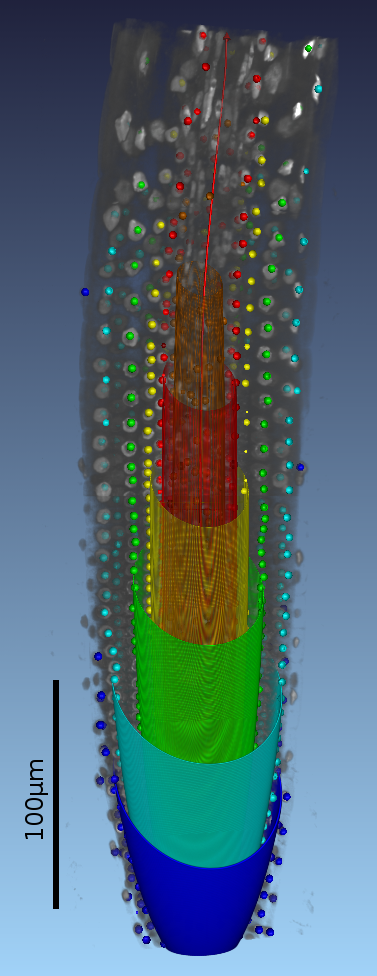
IntroductionArabidopis is an important model organism in molecular plant sciences. To understand various signalling pathways precise measurements of growth rates of responsive organs are essential [2]. GoalsThe goal of this work is to automate the measurement of photoreceptor controlled hypocotyl growth rates with computer vision techniques. ApproachOn a 2D image, the hypocotyl and the root can be delineated by two almost parallel curves. This makes the Lateral Coupled Snake (LCS) model [1] a suitable tool for analyzing seedling videos. The LCS model not only gives the contour of a seedling but also defines at every position the longitudinal direction of the seedling stem. When the starting and end points are located, the length of the hypocotyl is ready to be obtained from the model. AchievementsPositions and shapes of seedlings over time are well located and described by the LCS model. An example of the current result is shown in the following figure, from which one can find that the model acts like a ruler measuring length along a seedling. This work can be extended to root analysis as well. |
|
|
References
|
Analysis of Organ Development On Sub-Cellular Resolution at the example of Arabidopsis Thaliana
Members: Thorsten Schmidt, Junior-Prof. Dr. Olaf Ronneberger, Prof. Dr.-Ing. Hans Burkhardt
Partners: Dr. Taras Pasternak, Dr. Alexander Dovzhenko (Department of Botany, University of Freiburg), Prof. Dr. Klaus Palme
IntroductionIn contrast to single cell and cell culture approaches, the goal of this project is the description and modeling of signaling pathways between cells and tissues within their natural environment, e.g. the organism they form, given high resolution 3-D(+t) confocal recordings. GoalsUsing the example of the Arabidopsis Root Apical Meristem, we are going to provide tools for the statistical analysis of cellular key events within a complex organ with sub-cellular resolution. The approach will allow comparative studies within and between different plant populations after genetic manipulations, application of external stress or drug/hormone treatments. ApproachThe analysis and modeling consists of several steps:
AchievementsWe are able to automatically extract the positions of the nuclei and roughly estimate their nucleolus' diameter employing vectorial gray-value based invariants. In a supervised training/classification cycle we are able to distinguish Interphase nuclei from Mitoses and are now aiming at refining the model to describe the distributions of key events in relation to the root anatomy. Publications
|
|
Analysis of Trichome Patterning and Leaf Anatomy of Arabidopsis Thaliana in 4D Confocal Datasets
Members: Robert Bensch, Junior-Prof. Dr. Olaf Ronneberger, Prof. Dr.-Ing. Hans Burkhardt
Partners: Bettina Greese, Christian Fleck (Center for Biological Systems Analysis (ZBSA), Faculty of Biology, University of Freiburg), Katja Wester, Martin Hülskamp (Botanical Institute III, University of Köln)
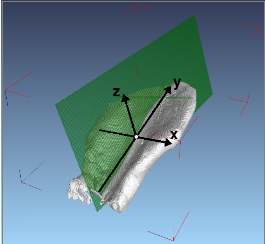
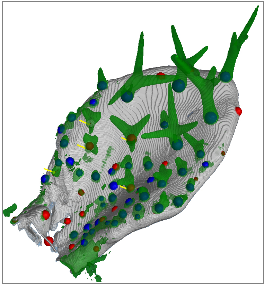
IntroductionArabidopsis trichomes (leaf hairs) are an interesting example to study cell differentiation [reference 1] and are well suited cell types for the functional analysis of de novo pattern formation. During cell growth, different processes provide for a regular spacing of the trichomes. A close interplay between theoretical modelling and experimental validations has been started to test the relevance of different models for trichome patterning. In this context, there is a need of quantitative data, e.g. trichome positions, to allow the analysis of these processes and their simulation with mathematical models. GoalsGoal of this project is the automated extraction of quantitative information about trichome patterning on leaves of Arabidopsis thaliana. The extracted quantitative data, e.g. trichome positions, are to support the analysis of these processes and their simulation with mathematical models. ApproachFor this work time series of growing rosette leaves (4D confocal datasets, 3D + time) are used. The approach mainly consists of three parts. The first part is the robust extraction of the leaf surface and midplane from the chlorophyll channel. Local filtering based on gradient directions and gradient magnitude is used to identify surface voxels. In addition voxels belonging to the midplane are identified similarly by using eigenvectors of the Hessian instead. Robustness is increased by applying a scale-space approach and removing unstable responses. The consideration of 4D data in our approach introduces the need for image registration. Symmetry analysis, based on the extracted surface, yields the symmetry plane of the leaf. It is used to robustly detect the pose of the leaf and thus to register a biologically motivated reference coordinate system. This allows for inter-subject registration of the leaves. Trichomes are localized in the GFP channel by first detecting candidates using Hough transform, extracting local 3D invariants, and then validating the candidates using a SVM.
Achievements
References
Publications
|
|
This page is maintained by the project responsibles.



