Open Source Software
| Binaries/Code | Datasets | Open Source Software |
| libsvmtl | ImageJ-Plugins | XuvTools | Presto-Box | iRoCS | U-Net |
U-Net
Source Code
We provide source code for caffe that allows to train U-Nets (Ronneberger et al., 2015) with image data (2D) as well as volumetric data (3D). The code is an extension to the previously released work that implemented 2D U-Nets. The patch contained in caffe_unet_3D_v1.0.tar.gz implements the layers for 2D and 3D U-Net including the value augmentation and random elastic deformation. If you use this code, please cite (Ronneberger et al., 2015) and/or (Çiçek, et al., 2016). Although the software does not require a GPU (i.e. can be run on the CPU only), we strongly recommend an NVIDIA GPU with compute capability >= 3.0. Installation is supported for Linux and Mac. The procedure and dependencies are the same as for the installation of the original BVLC/master branch of caffe. Installation instructions are available for different Linux distributions and for Mac. See the accompanying LICENSE file for copyright notice. Future updates and bugfixes will be published on this site. The README file contains installation and usage notes.Models and Examples
- In the experiments of our MICCAI 2016 paper, we have used two types of network architectures: without batch normalization, with batch normalization. In order to reproduce our results you can see solver parameters we chose for all our experiments. After the successful installation and the architectural choice, you can start training your 3D U-Net with this example command. Here you can find an example of how the trainfileList.txt should look like. In order to test your trained models, we provide the matlab script 3d_unet_predict.m which performs testing.
- Hints about input and output: Both input and output should be 5D blobs arranged as (#of samples, #of channels, depth, height, width). Note that the annotations should be single channel data including integer labels per pixel. If you are running our model on your data, random_offset_from and random_offset_to parameters in the create_deformation_layer must be changed since they are the only data size dependent parameters in our model. See findsize.m to see how to compute them properly together with input and output sizes for our 3D u-net given a specific bottom blob shape.
- For a trained 3D U-Net model (corresponds to the experiment reported in first column and first row of Table 1 in 3D U-Net paper) please see example caffemodel.
How to Cite
- If you plan to publish work that uses or incorporates our contributions in the code provided here, please cite our work:
Olaf Ronneberger, Philipp Fischer & Thomas Brox. U-Net: Convolutional Networks for Biomedical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol.9351, 234--241, 2015  Publisher's Link
Publisher's Link
 Code
Code
Özgün Çiçek, Ahmed Abdulkadir, S. Lienkamp, Thomas Brox & Olaf Ronneberger. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol.9901, 424--432, Oct 2016 
 Publisher's Link
Publisher's Link
 Code
Code
- Other related work:
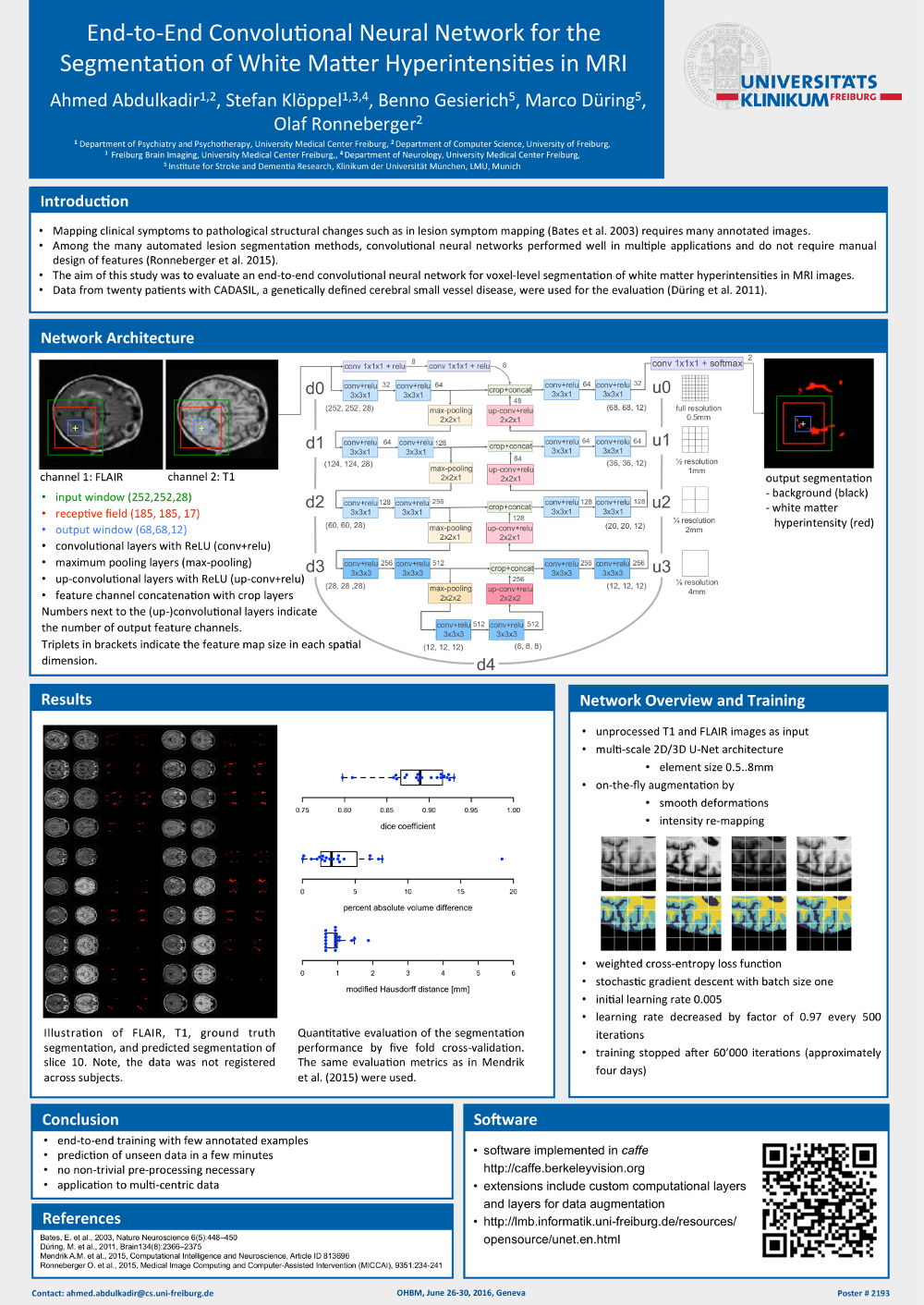
Ahmed Abdulkadir, Stefan Klöppel, Benno Gesierich, Marco Düring & Olaf Ronneberger. End-to-End Convolutional Neural Network for the Segmentation of White Matter Hyperintensities in MRI. 22nd Annual Meeting of the Organization for Human Brain Mapping (OHBM), Geneva, Switzerland, 2016 (Poster presentation)
Contact: Ahmed Abdulkadir, Robert Bensch, Özgün Çiçek, Thorsten Falk, Olaf Ronneberger


